Some repositories & Code showcase
paginate pandas (Interactive pagination for pandas.DataFrames)
https://github.com/jmberros/paginate_pandas
paginate pandas is a dead-simple interactive paginator for a pandas Series or DataFrames that leverages on ipywidgets and works beautifully on Jupyter Lab. It let’s you easily browse through a possibly big (say, hundreds of thousands of rows) dataframe advancing page by page. It also let’s you control the page size. If you deal with pandas a lot in Jupyter, this will change your quality of (developer) life.
I developed it out of the necessity of exploring large tables from Jupyter.
paip (DNA data analysis pipelines)
https://github.com/biocodices/paip
paip is a Luigi-based Python pipeline for the analysis of DNA sequencing NGS data. It has routines for quality control, variant calling and report generation, with a comprehensive unit test suite for pytest. I coded this in the span of 2 years as new needs arised in a medical genomics laboratory.
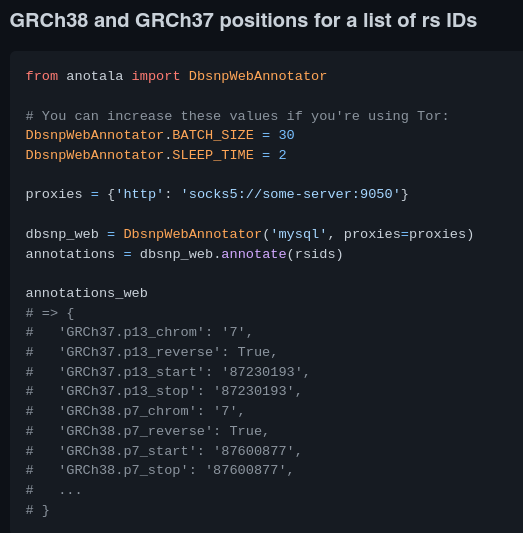
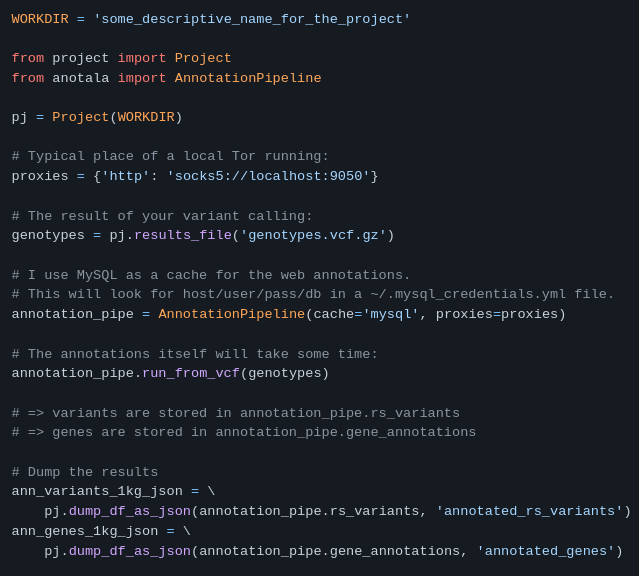
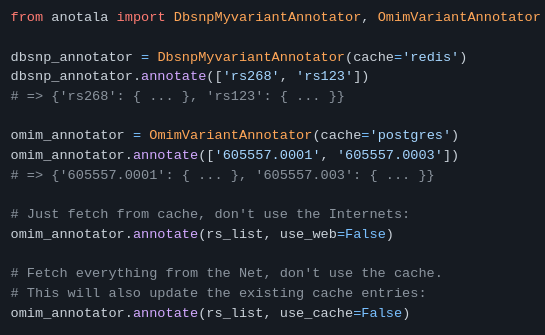
anotala (annotation of mutations)
https://github.com/biocodices/anotala
anotala is an Python bioinformatics utilty that lets you annotate DNA mutations, i.e. add extra information to mutations found in a sample like aminoacid/protein affected, coding vs. non-coding and other region types, predicted strength of effect with different softwares, diseases known to be associated with the mutation.
It implements a cache that can be based on MySQL, PostgreSQL or Redis.
More use cases in the repo’s README.
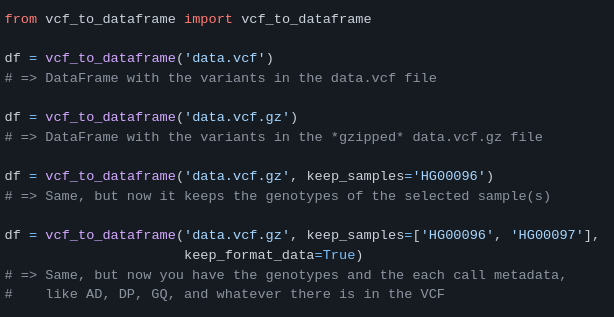
vcf to dataframe (DNA data format conversion)
https://github.com/biocodices/vcf_to_dataframe
vcf_to_dataframe is a Python utilty to import a VCF file (a standard format to store DNA variants) as a pandas DataFrame. It allows to import any subset of samples in the file and it converts all INFO and GT fields to Python lists or dicts to ease the downstream parsing.
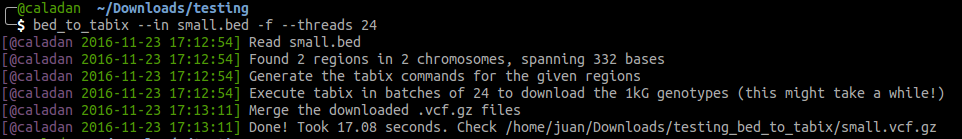
bed to tabix (1KGP genotypes downloader)
https://github.com/biocodices/bed_to_tabix
bed_to-tabix is a friendly Python CLI utility that will download the genotypes of 2,504 individuals of the 1,000 Genomes Project (1KGP) for the genome regions define in an input BED file. The script deals with chromosome separate downloading and merging, and with temporary files clenaup.