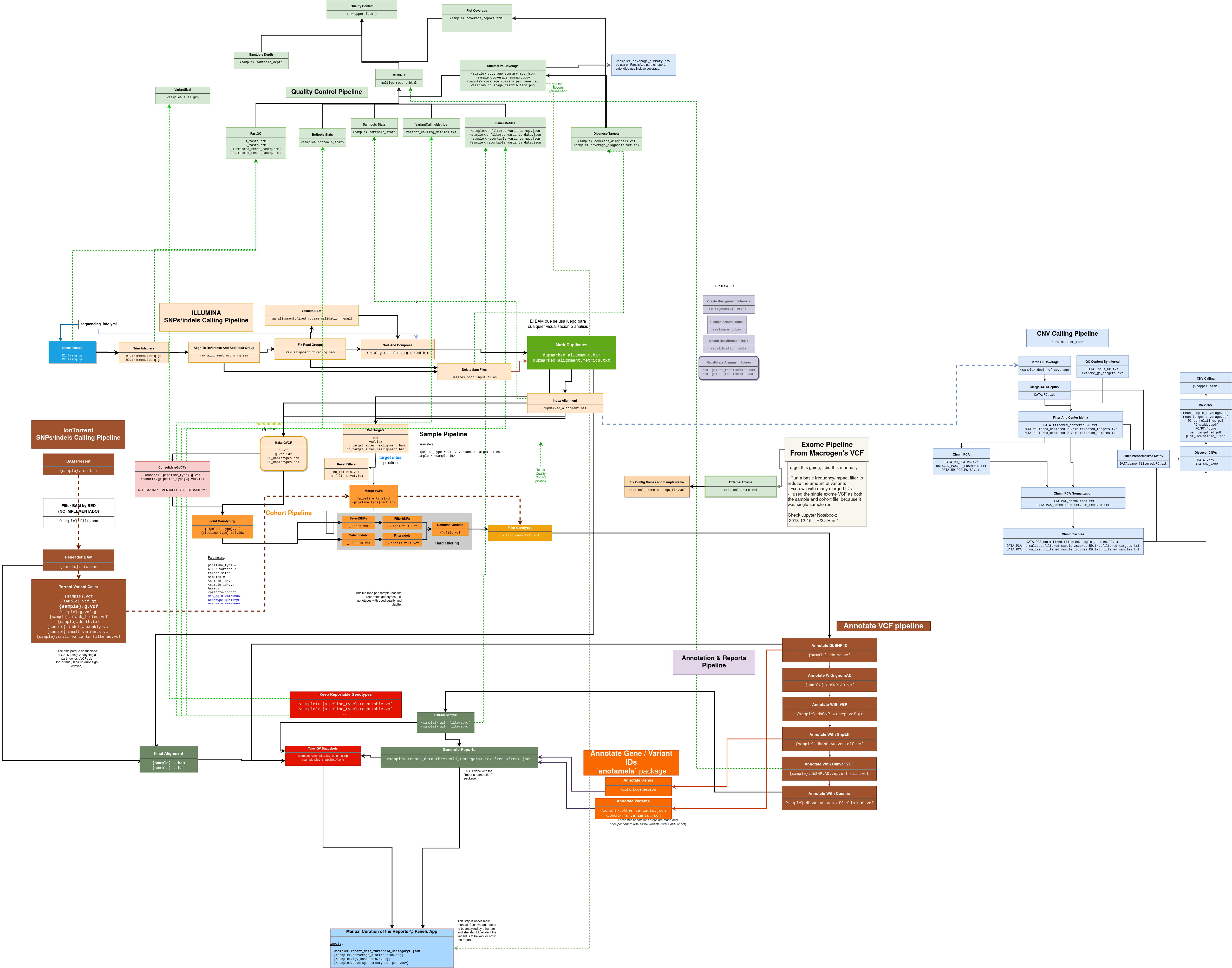
Complex Pipeline of Genetic Data Processing
In the Biocodices Medical Genetics laboratory, I built a complex pipeline of genetic data processing based on the Python’s workflow framework Luigi (a framework similar to Airflow, developed at Spotify).
The workflow started with the raw output of various DNA Sequencers (i.e. machines that read the sequence of DNA samples) and went through numerous standard bioinformatic quality assurance and filtering steps until a subset of medically relevant genetic mutations was kept for reporting.
Multiple workflows were created around the main “mutations detection” workflow:
- The main small mutations detection workflow, based on Illumina NGS data.
- A specific workflow for data of a different sequencer called IonTorrent.
- A side workflow of data quality analysis of partial results at various steps of the main pipeline.
- An ever-growing “annotation” pipeline to provide additional information about each detected mutation, feeding from multiple medical and population genetics sources.
Multiple filetypes were manipulated by the pipeline: JSON, PNG images, bioinformatic formats like VCF, BAM, FASTQ, and even the automatic generation of a software-specific script to generate genome “screenshots”.
Big efforts had to be done not to overcrowd the servers with intermediate files, while keeping a track of the intermediate steps for debugging and quality analysis.
The full pipeline is shown in the following graph for illustration purposes:
The code is open and can be explored in the Github repo.